1.
Orang Asli (Malaysian Itas: Kensui and Jehai)
2.
Ati Ita (Philippine Itas: Ayta, Ati, Agta, &
Iraya, North Ita)
3.
Mamanwa Ita (Philippine Ita, South Ita)
4.
Papuan
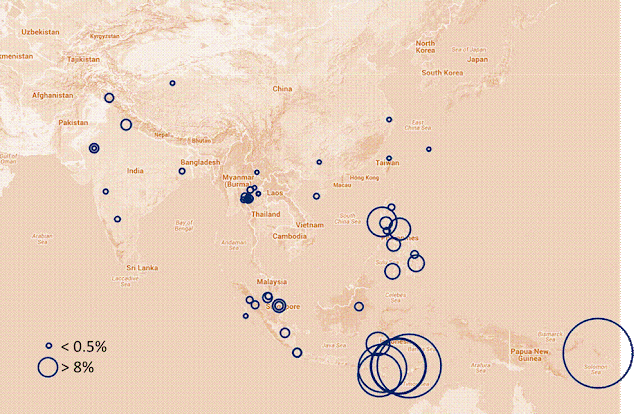
The same can be said with the separation for the two groups
of Itas in the Philippines, the Mamanwa and North Itas. We can probably deduce
from this that the separation of the two Philippine Ita populations was due to
at least two major island groups that formed the Philippines during those ice
ages. If we look carefully at the Philippine map including the seabed, you can
almost make out at least two island groups. The Luzon – North Bisaya island and
the South Bisaya - Mindanao island. Again, these long period of physical
separation probably also caused the large genetic distance.
It’s interesting that the Papuans and Atis are close to each
other; I would have expected the Atis and Mamanwas would be closer due to
proximity. Perhaps the Mamanwas had a “bottle neck” founder populations similar
to the Mlabri, meaning, the founders of the Mamanwa before the separation came
from a small group of the bigger Ita group (bottle neck).
Figure 6: Mamanwa Gene Prevalence
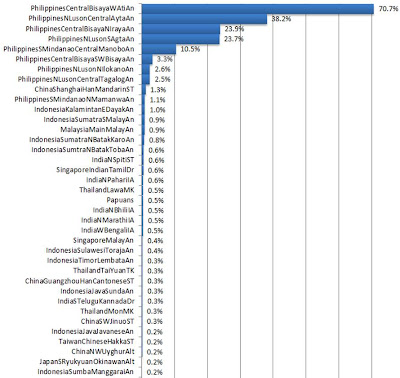
Figure 7: Ati Gene Percentage
Figure 8: Ati Gene
Prevalence
Figure 9
& Figure 10
bring in some inferences. The Ati & Mamanwa groups have only recently
admixed with the Malay speakers (Nusantao). This is likely since the
archaeological evidence of Malay settlements to ISEA are less than 5.5K BP. The
significant Papuan presence in the Ati samples is a surprise. As mentioned, the
four Ita groups have large separation. If it is true then the Papuan content is
of recent admixture. Were the Papuans and/or Atis able to develop maritime
technology (sea worthy ships)? The other possibility is that the Ati’s were the
founder source for the Papuans (and perhaps the Mamanwas); given the relatively
greater Ati diversity. Note the Mamanwas has 1.1% Papuan content. The Papuans,
on the other hand, has 99.2% Papuan, 0.5% Ati, & 0.3% Mamanwa. Whatever the
possibilities are, the Papuans, Mamanwas & Atis have significant
interactions in Luson, Bisaya, & Mindanao.
Note: Although percentage less than 0.25% maybe be a margin
of error in admixture analysis, I would not completely dismiss these results
since we are only comparing 55,000 SNPs compared to the millions unidentified.
For the moment, I will most likely not explore percentage less than 0.25%.
Figure 9: Mamanwa Admixture
Figure 10: Ati Admixture
Figure 11:
Papuan Gene Percentage
Figure 12:
Papuan Gene Prevalence
Figure 13:
Malaysia Ita Gene Prevalence
Figure 14:
Malaysian Ita Admixture
References
1. Yang X, Xu
S, The HUGO Pan-Asian SNP Consortium (2011) Identification of Close Relatives
in the HUGO Pan-Asian SNP Database. PLoS ONE 6(12): e29502.
doi:10.1371/journal.pone.0029502
2. D.H.
Alexander, J. Novembre, and K. Lange. Fast model-based estimation of ancestry
in unrelated individuals. Genome Research, 19:1655–1664, 2009
3. H. Zhou,
D. H. Alexander, and K. Lange. A quasi-Newton method for accelerating the
convergence of iterative optimization algorithms. Statistics and Computing,
2009.
4. Alexander
D. H., Lange K. (2011). Enhancements to the ADMIXTURE algorithm for individual
ancestry estimation. BMC Bioinformatics 12:246. doi: 10.1186/1471-2105-12-246.
5. Purcell S,
Neale B, Todd-Brown K, Thomas L, Ferreira MAR, Bender D, Maller J, Sklar P, de
Bakker PIW, Daly MJ & Sham PC (2007) PLINK: a toolset for whole-genome
association and population-based linkage analysis. American Journal of Human
Genetics, 81.
6. Greenhill,
S.J., Blust. R, & Gray, R.D. (2008). The Austronesian Basic Vocabulary
Database: From Bioinformatics to Lexomics. Evolutionary Bioinformatics,
4:271-283.
7. Mijares,
A.S.B. et al. 2010. New evidence for a 67,000-year-old human presence at Callao
Cave , Luzon , Philippines. Journal of Human Evolution, 59:123-132.
doi:10.1016/j.jhevol.2010.04.008.
8. Mijares,
A.S.B.2007. The Late Pleistocene to Early Holocene For-agers of Northern Luzon.
Bulletin of the Indo-Pacific Prehistoric Association 28:99-107.
9. Sagart, L.
(2002). Sino-Tibeto-Austronesian: An Updated and Improved Argument. BMC
Bioinformatics 12:246. doi: 10.1186/1471-2105-12-246.
10. Gaillard, J. C. and Mallari,
J. P. (2004), The peopling of the Philippines: A cartographic synthesis, Hukay:
Journal of the University of the Philippines Archaeological Studies Program 6.
11. Mijares, A.S.B. 2008. The
Peñablanca Flake Tools: An Unchanging Technology? Hukay 12:13-34.
12. Mijares, A.S.B. et al. 2010.
New evidence for a 67,000-year-old human presence at Callao Cave , Luzon ,
Philippines. Journal of Human Evolution, 59:123-132.
doi:10.1016/j.jhevol.2010.04.008.
13. Dizon, E.Z. et al. 2002.
Notes on the Morphology and Age of the Tabon Cave Fossil Homo Sapiens. Current
Anthropology 43:660- 666.
14. Détroit, F. 2002. Origine
et évolution des Homo sapiens en Asie du Sud-Est: Descriptions et analyses
morphomé-triques de nouveaux fossiles. PhD thesis, Paris, France: Muséum
national d'Histoire naturelle.
15. Détroit, F. et al. 2004.
Upper Pleistocene Homo sapiens from Tabon cave (Palawan, the Philippines). Human
Paleontology and Prehistory 3:705–712.
16. Fox, R.B. 1970. The Tabon
Caves. Monograph of the National Museum of the Philippines. No. 1. Manila.
17. Barton, H., Piper, P.J.,
Rabett, R., and Reeds, I., 2009. Com-posite hunting technologies from the
Terminal Pleisto-cene and Early Holocene, Niah Cave, Borneo. Journal of
Archaeological Science 36:1708–1714.
18. Kaplan, M. R. et al. (2005).
Cosmogenic nuclide chronology of pre-last glacial maximum moraines at Lago
Buenos Aires, 468S, Argentina. Science Direct Quaternary Research 63
(2005) 301 – 315.
19. Kennedy, K. A. R. 1977. The
deep skull of Niah. AP 20:32-50.
20. Brothwell, D. R. 1960. Upper
Pleistocene human skull from Niah Caves, Sarawak. SMJ 9:323-349.
21. Lews, H et al. 2008. Terminal
Pleistocene to mid-Holocene occupation and an early cremation burial at Ille
Cave, Palawan, Philippines, Antiquity Volume: 82 Number: 316 Page:
318–335
22. Sieveking, G. de G. 1954.
Excavations at Gua Cha, Kelantan 1954. Part 1. FMJ 1 and 2:75-143.
23. Adi Haji Taha. 1985. The
re-excavation of the rockshelter of Gua Cha, Ulu Kelantan, West Malaysia. FMJ
30.
24. Zuraina Majid. ed. 1994. The
Excavation of Gua Gunung Runtuh. Malaysia: Department of Museums and
Antiquity.
25. Budhisampurno, S. 1985.
Kerangka manusia dari Bukit Kelambai Stabat, Sumatera Utara. Pertemuan
Ilmiah Arkeologi III, 955-984. Jakarta: Pusat Penelitian Arkeologi
Nasiona1.
26. Solheim, Wilhelm G.
Archaeology and culture in Southeast Asia : unraveling the Nusantao, (revised
edition), Diliman, Quezon City : University of the Philippines Press, 2006.
27. Purcell S, Neale B,
Todd-Brown K, Thomas L, Ferreira MAR, Bender D, Maller J, Sklar P, de Bakker
PIW, Daly MJ & Sham PC (2007) PLINK: a toolset for whole-genome association
and population-based linkage analysis. American Journal of Human Genetics, 81.